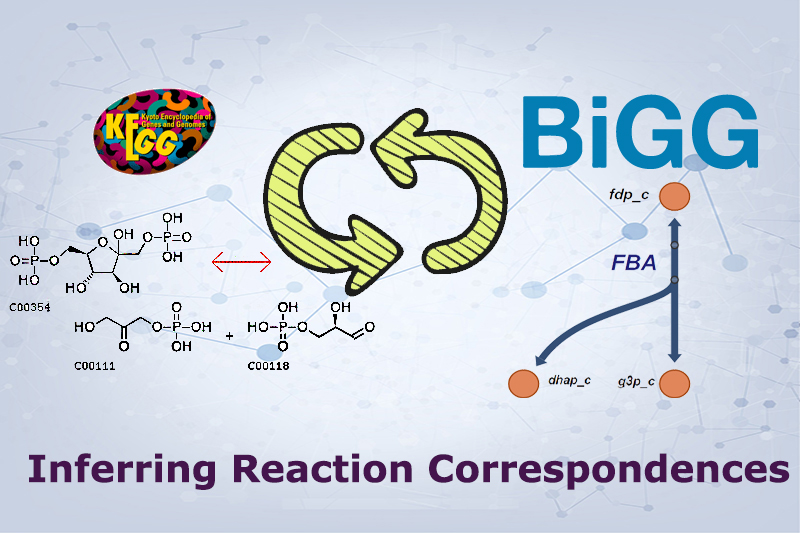
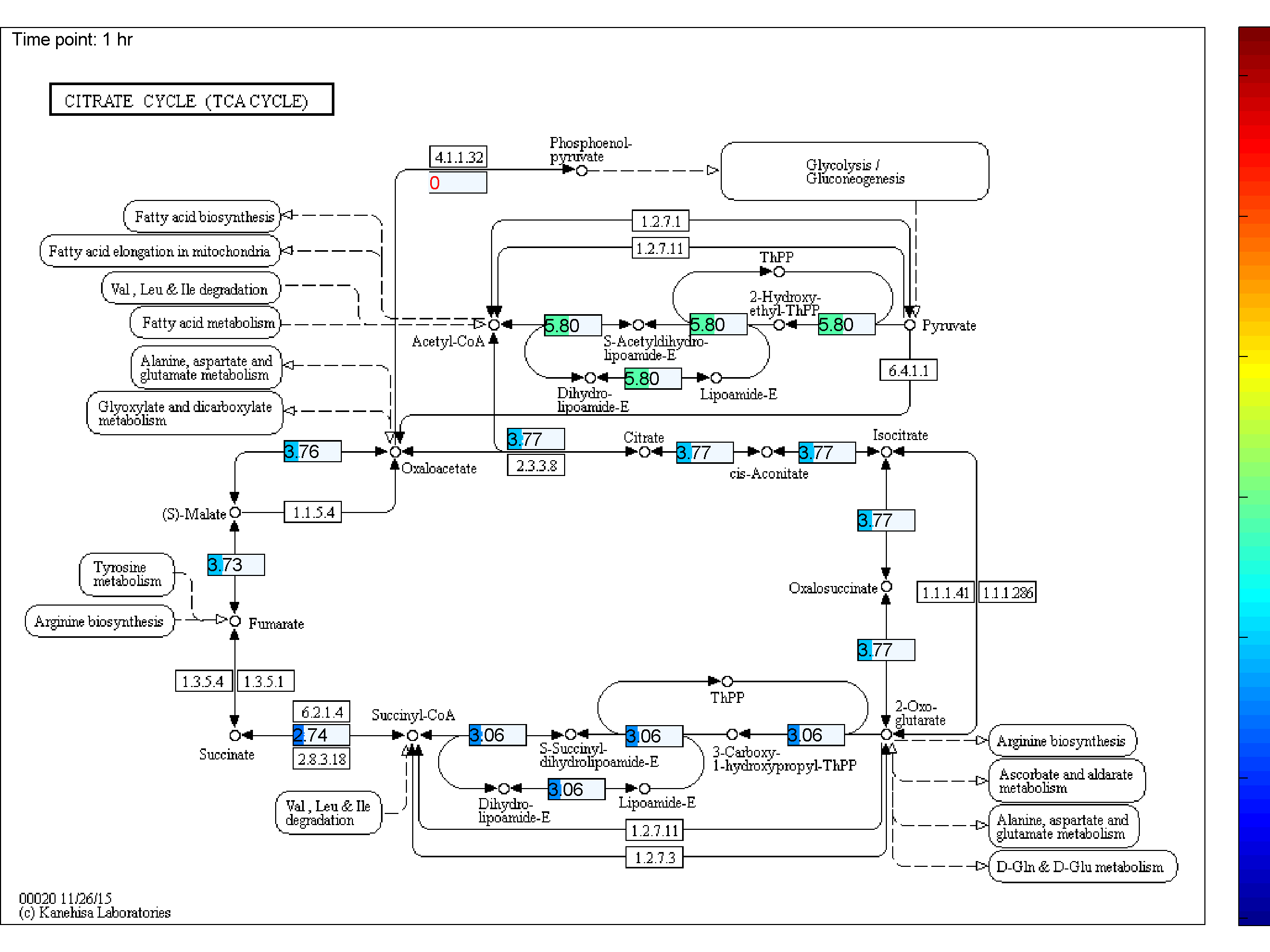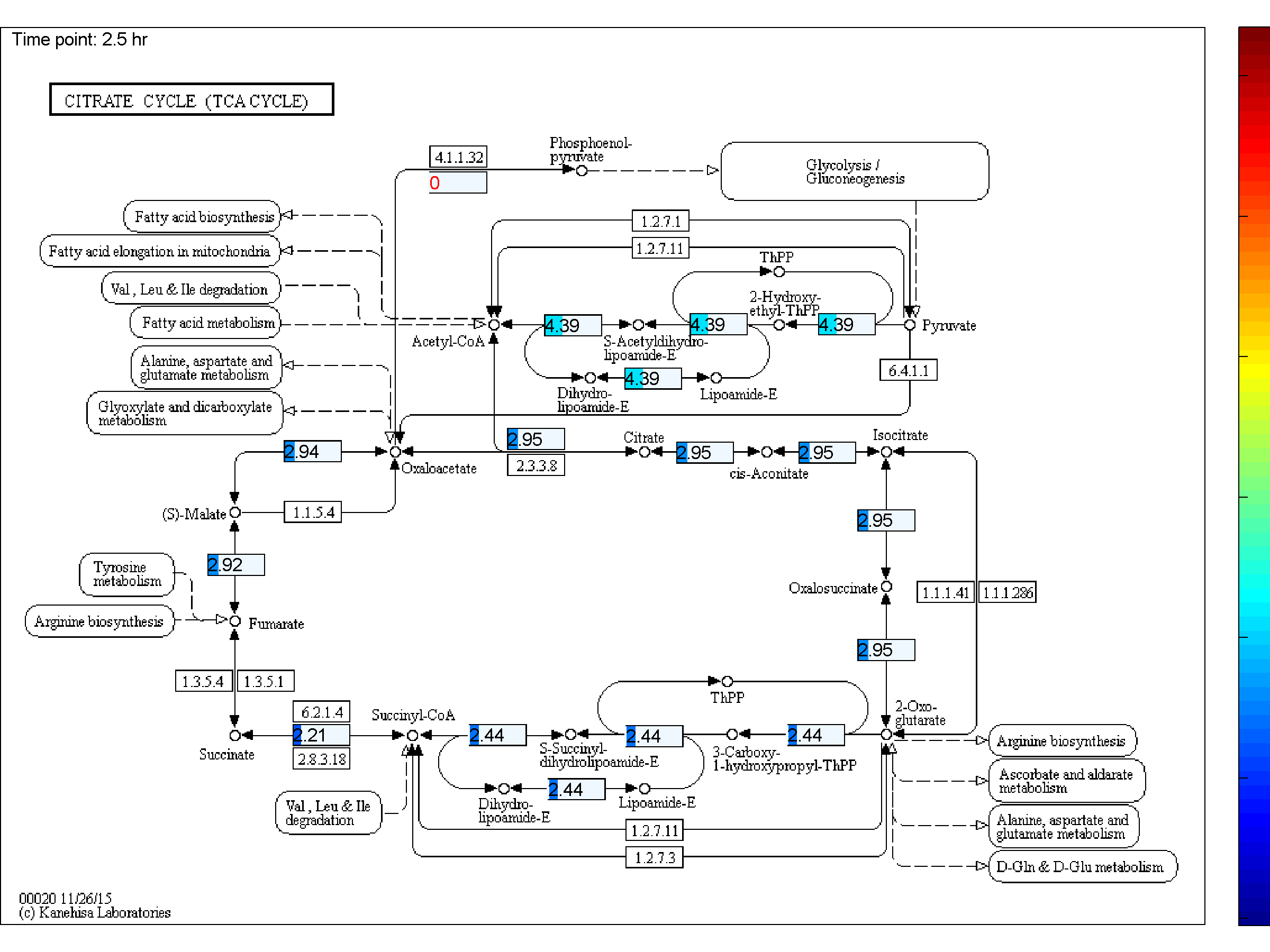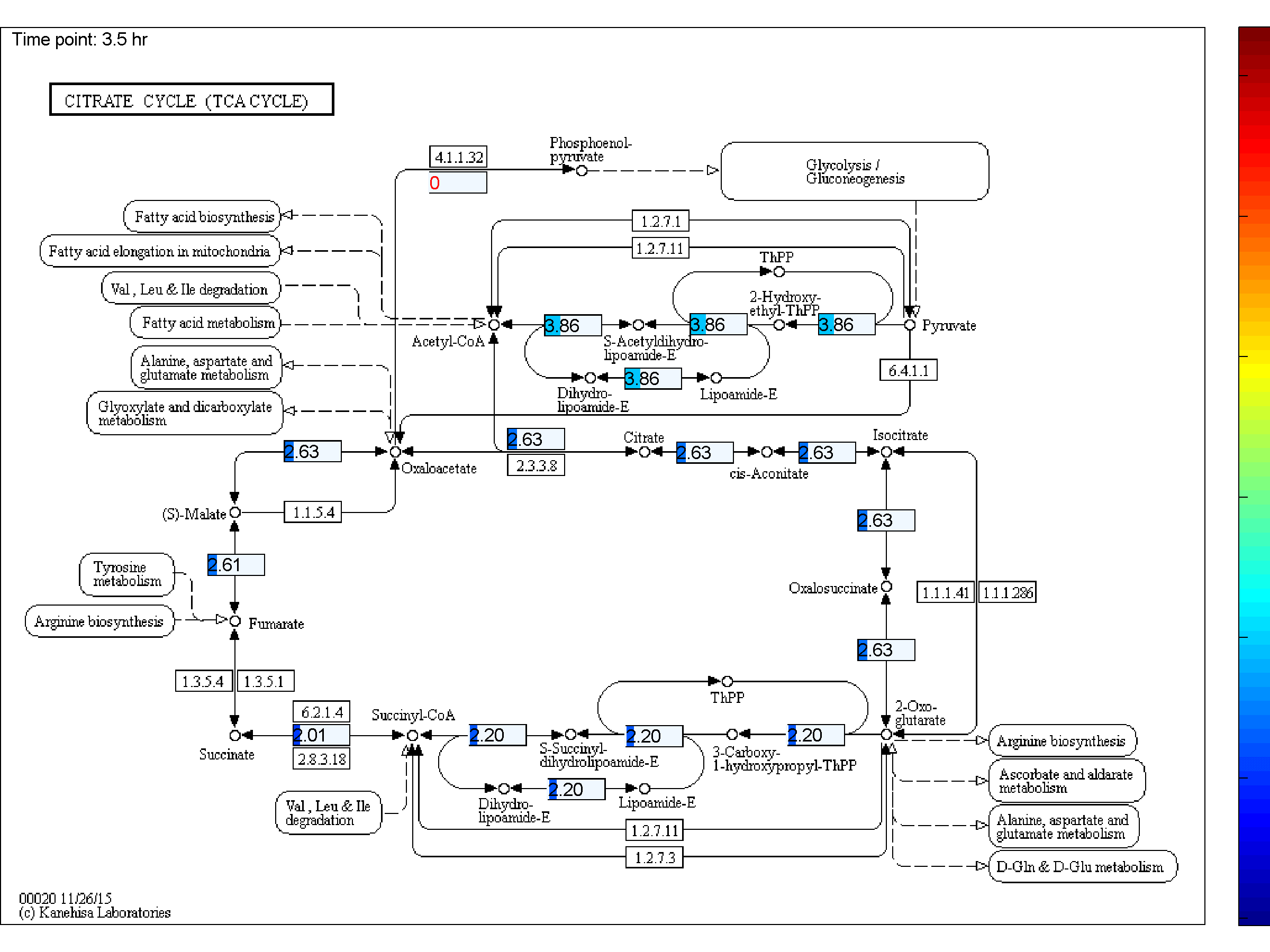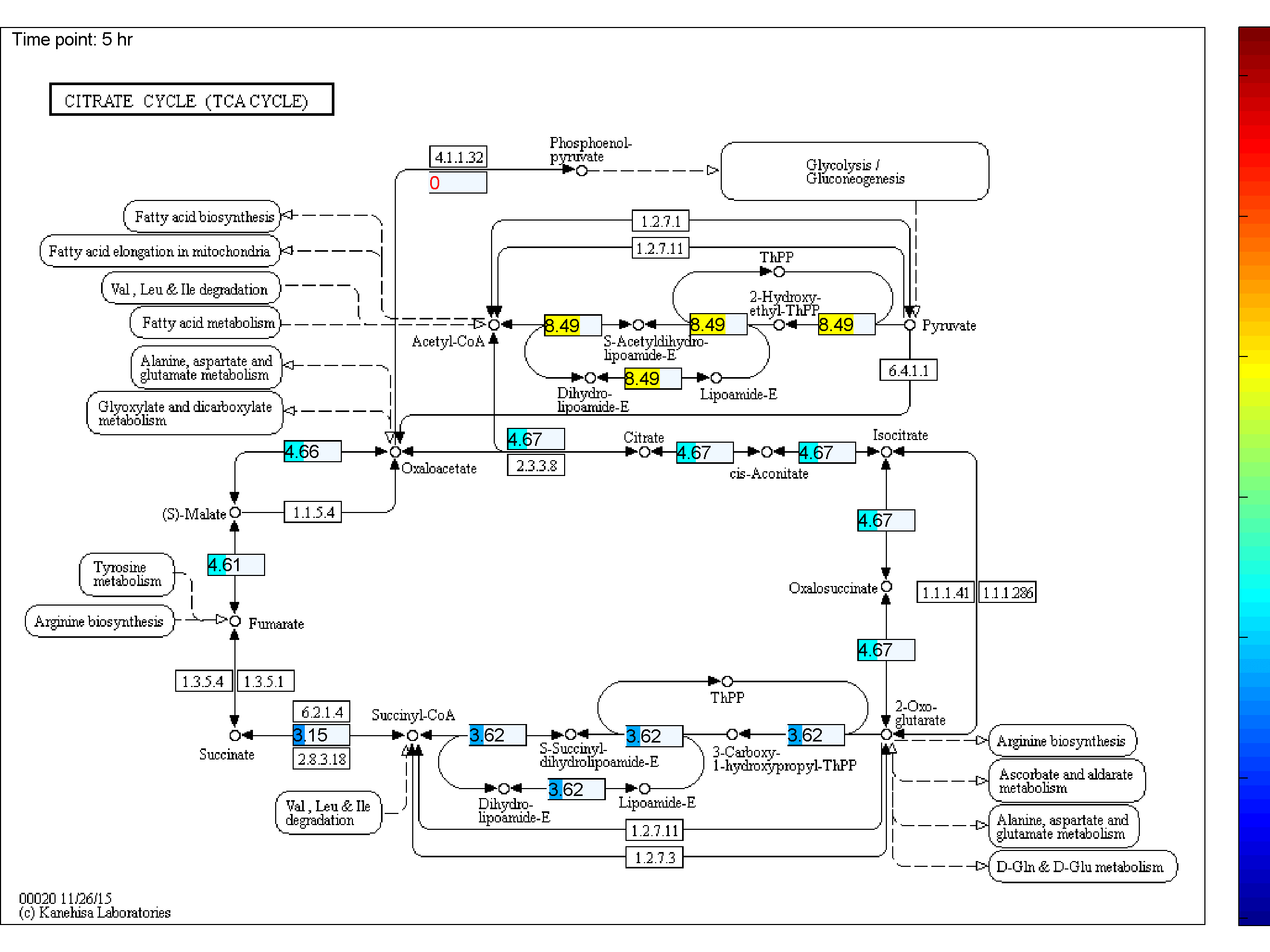
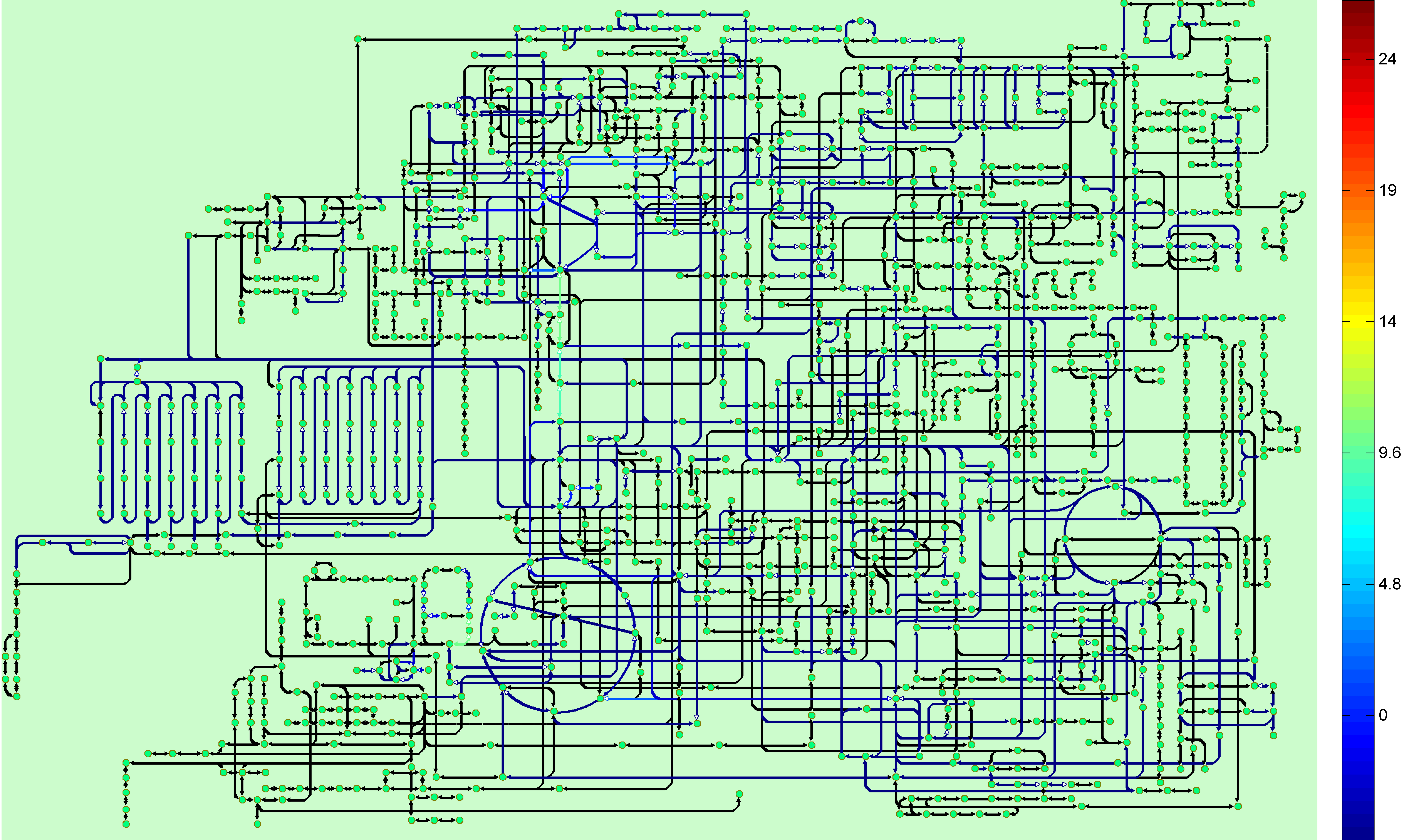
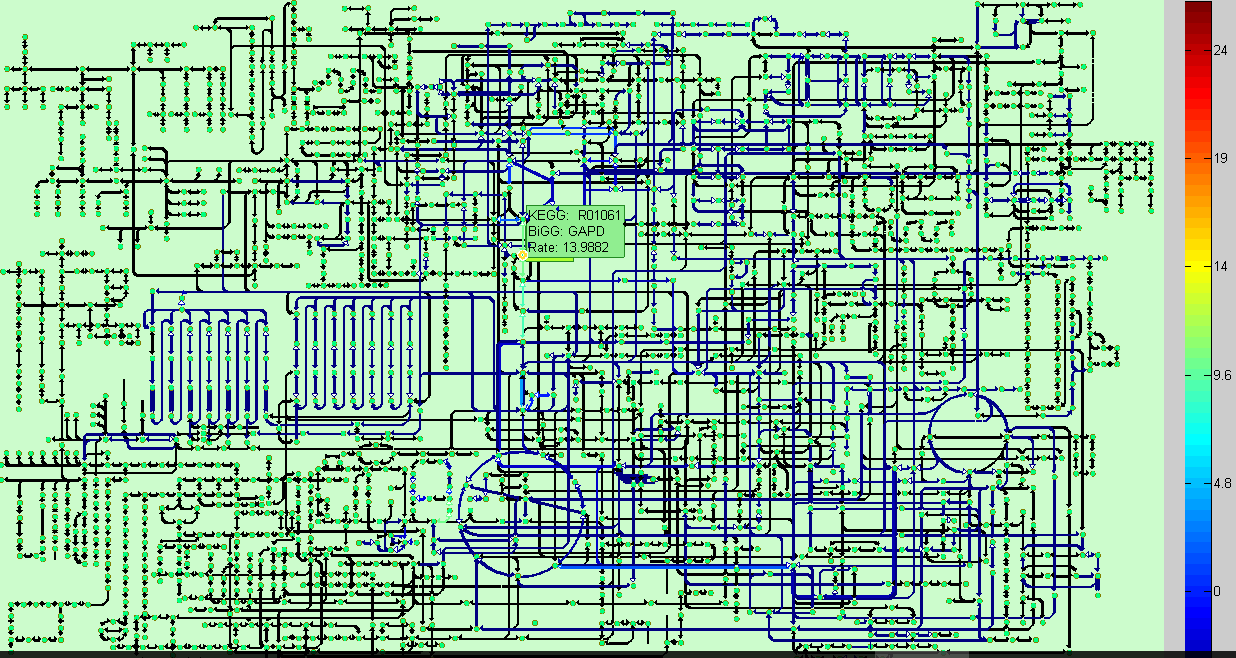
BiKEGG is an open source COBRA toolbox extension, which connects the KEGG and BiGG
databases by determining the reaction correspondences between reaction identifiers in
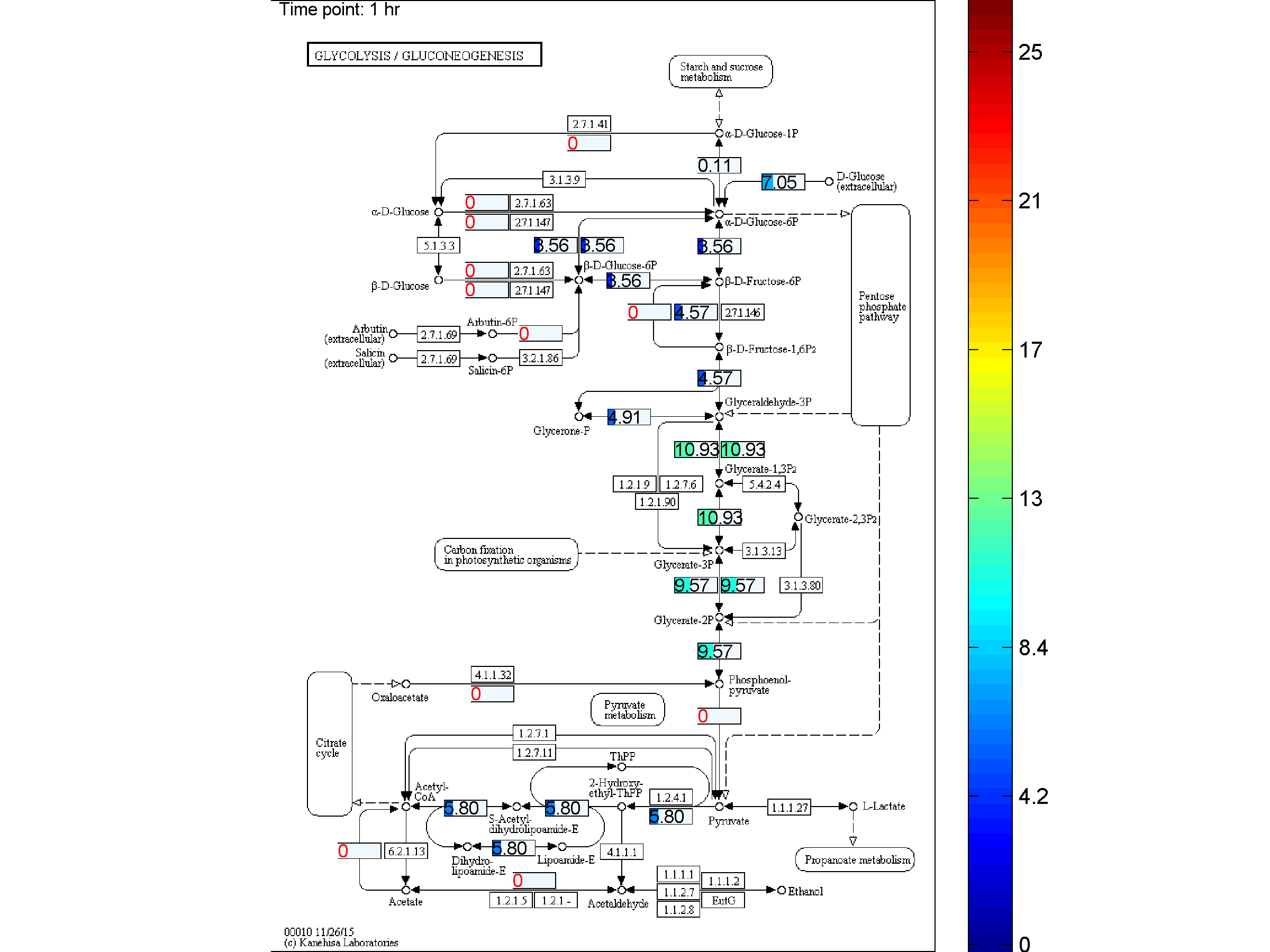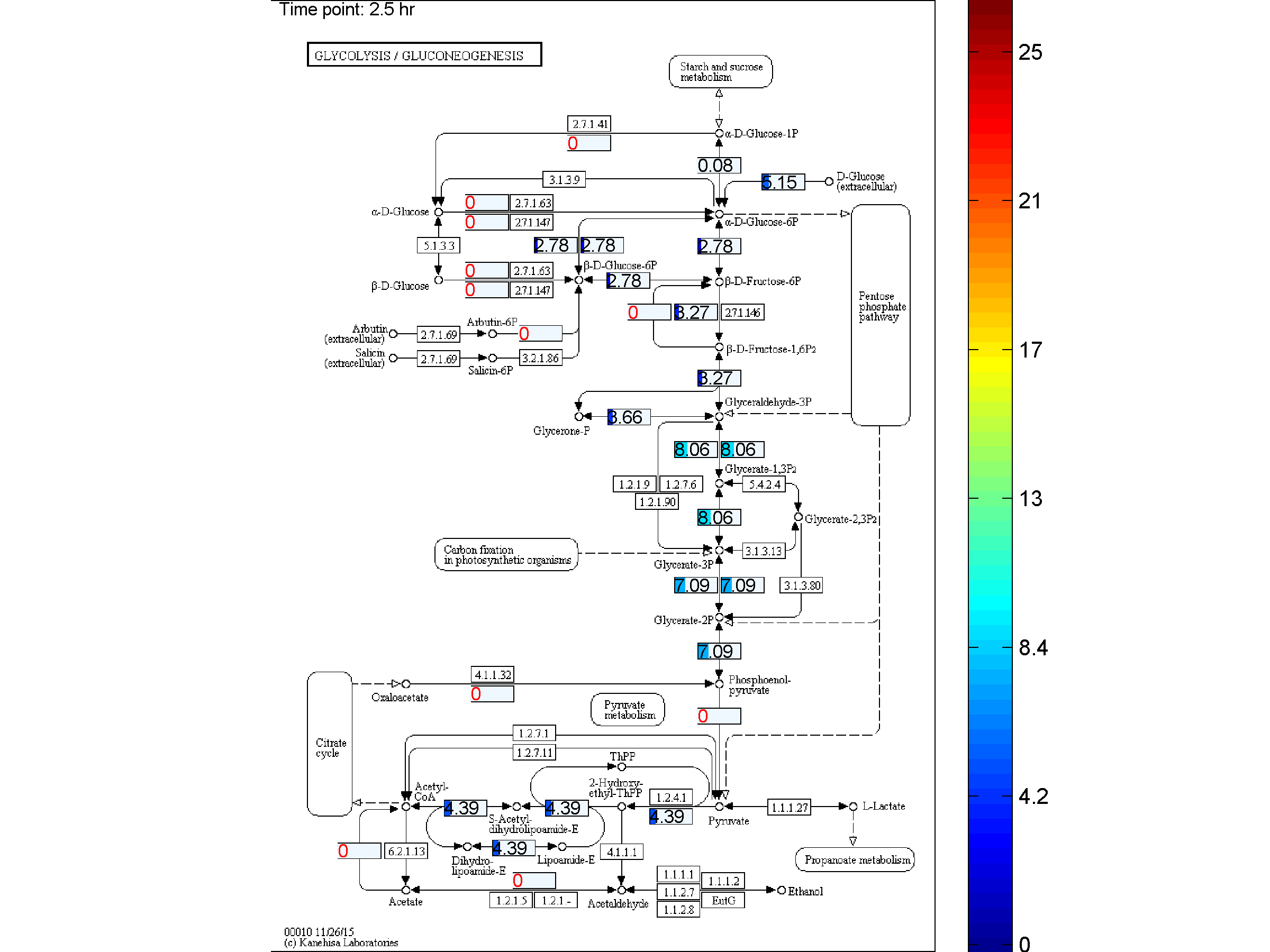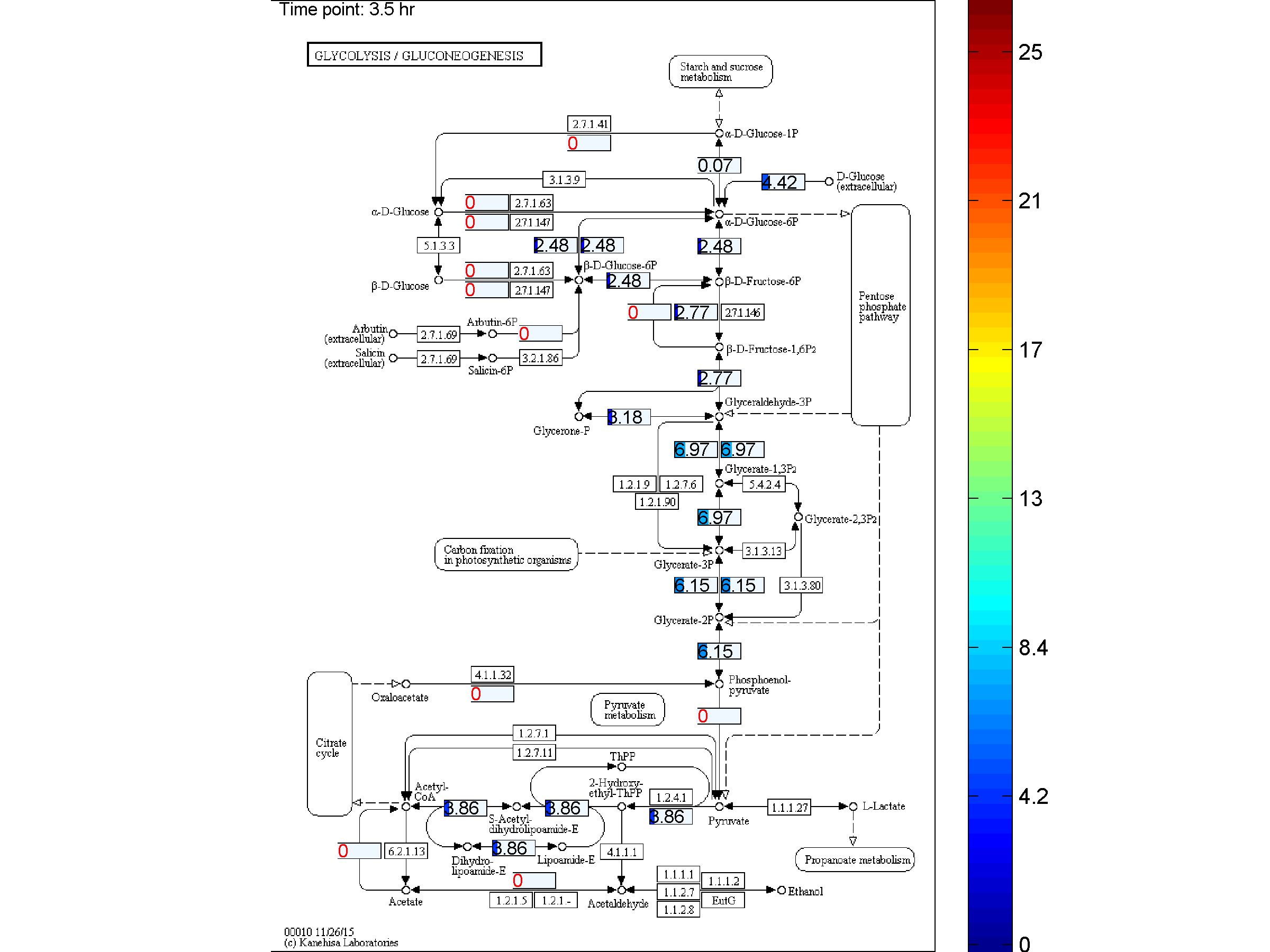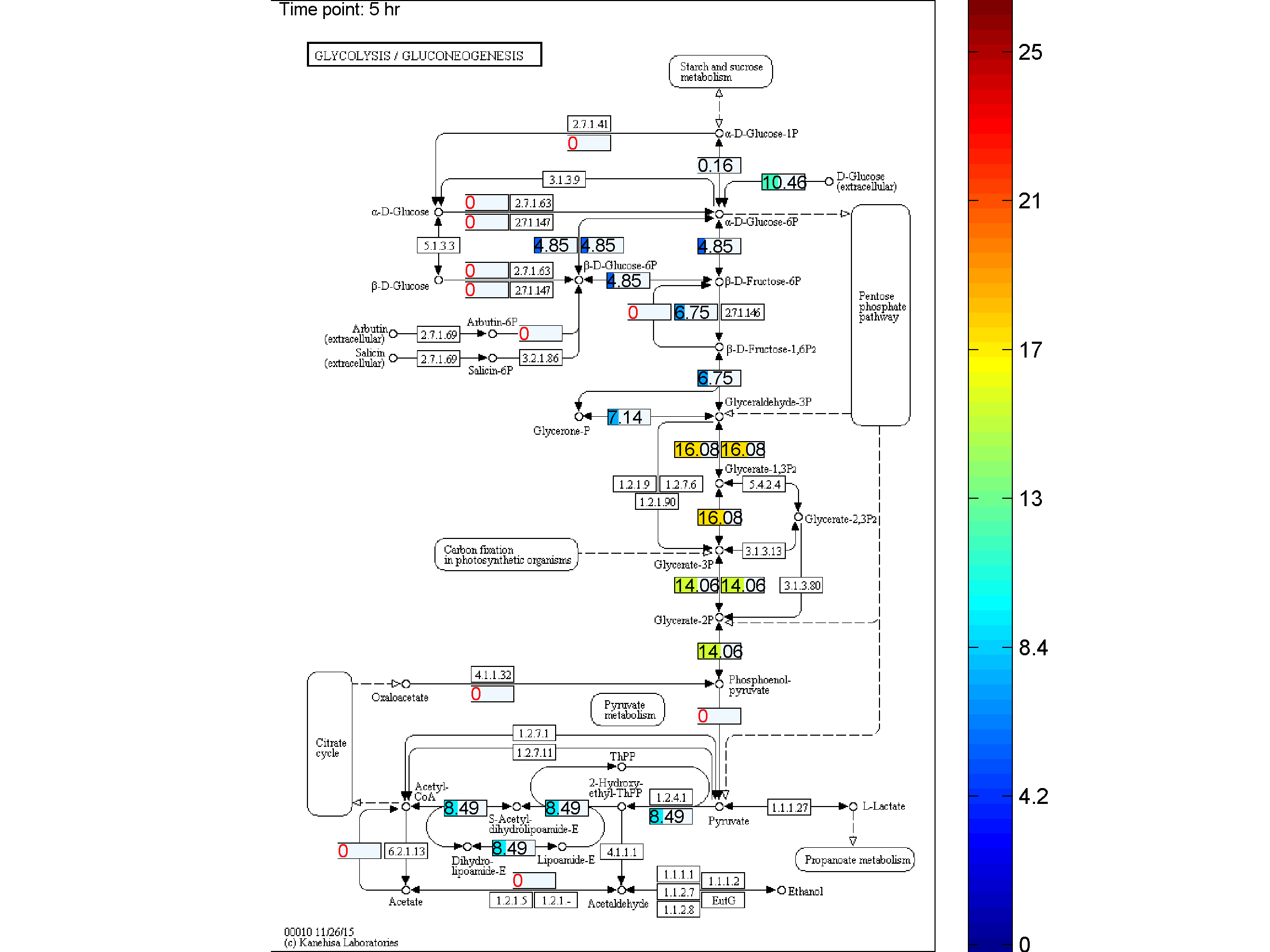
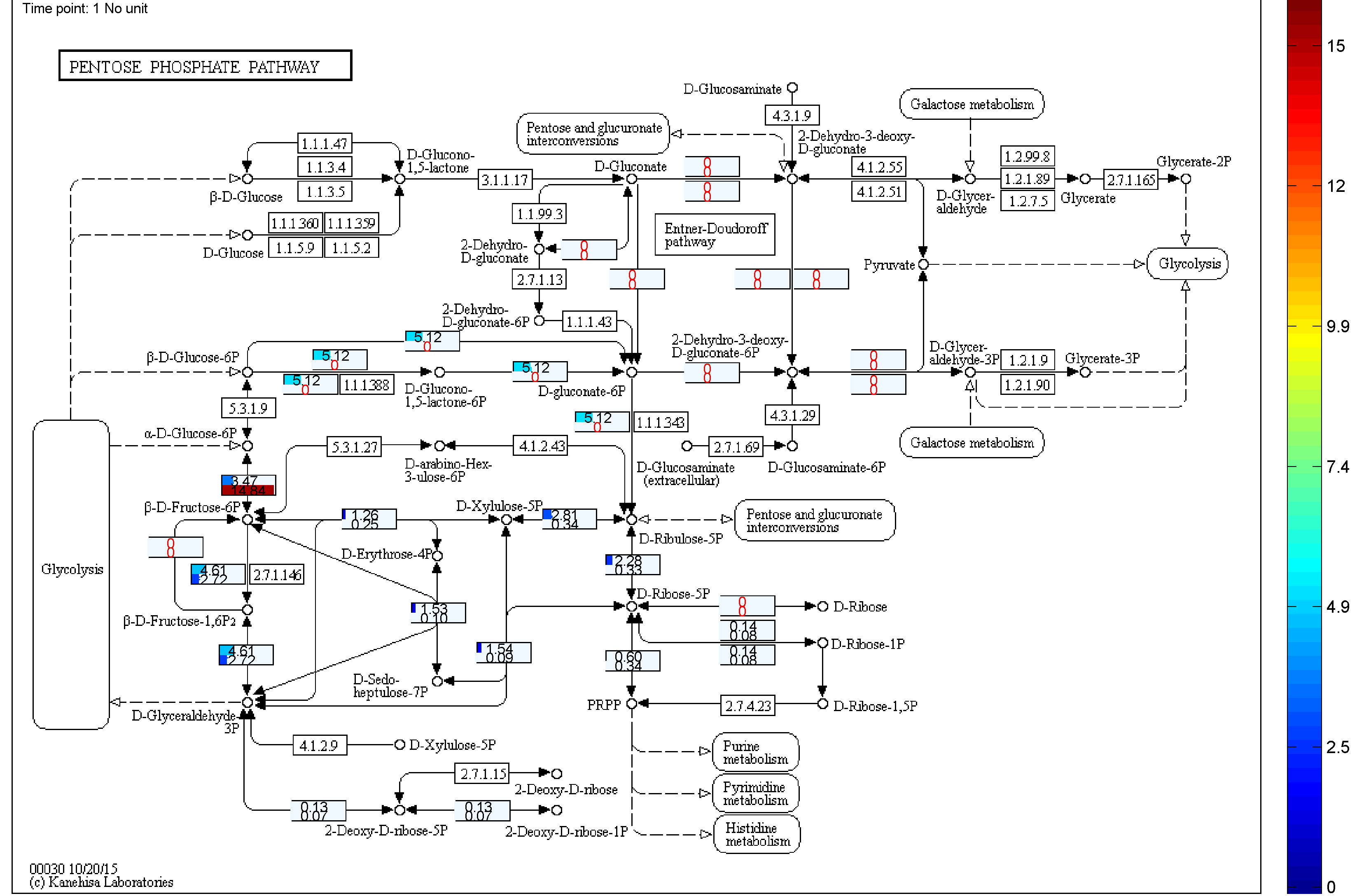
the two databases. BiKEGG is capable of overlaying the flux distributions on KEGG PATHWAY
maps and finding corresponding reactions for an organism of interest based on the genome
annotation presented in KEGG.
Cite: Jamialahmadi, O., Motamedian, E., Hashemi-Najafabadi, S. BiKEGG: A COBRA toolbox extension for bridging the BiGG and KEGG databases. Mol. BioSyst. 2016, DOI: 10.1039/C6MB00532B.
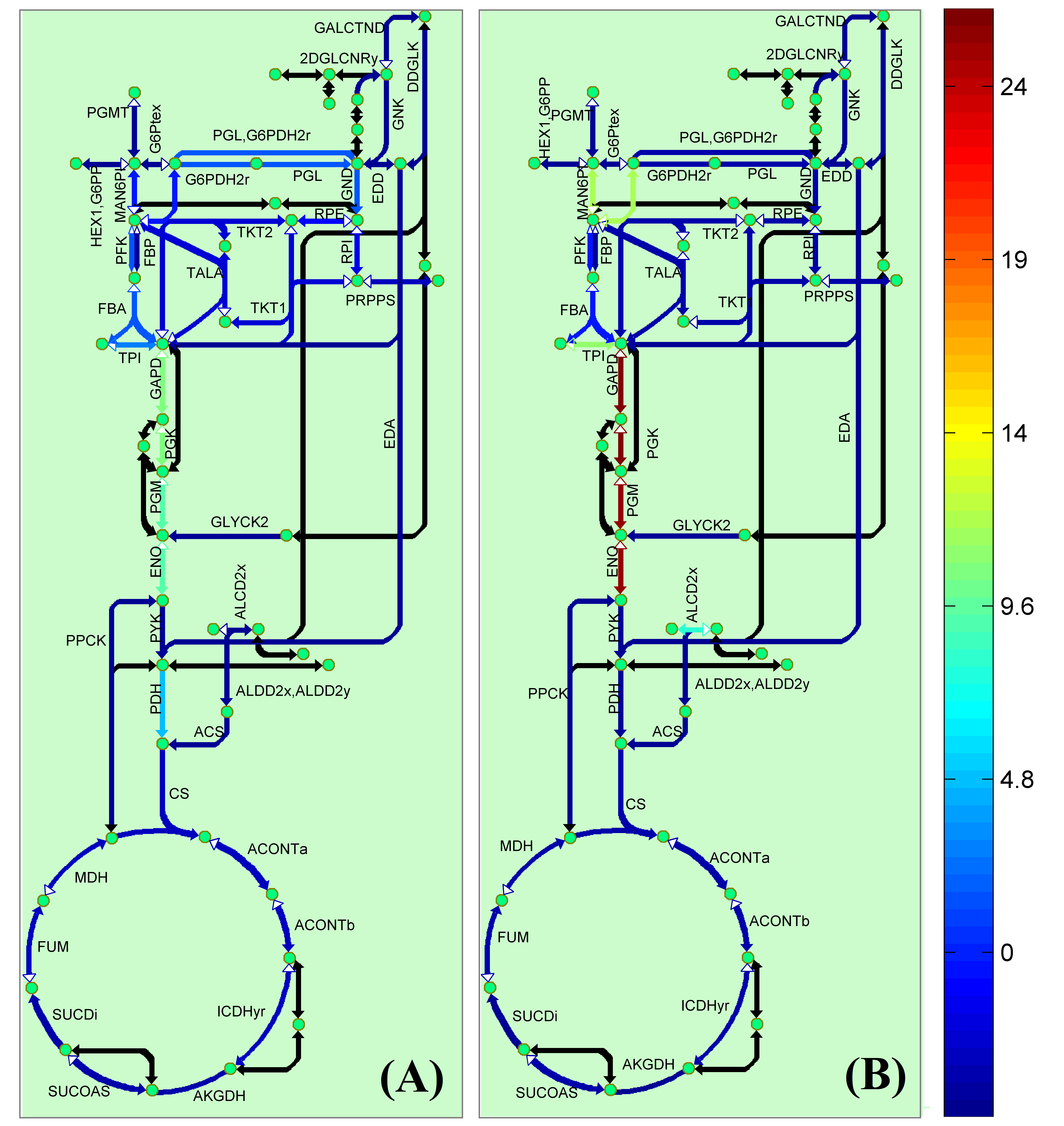
Visualization
Classical KEGG pathways | Customized metabolic maps
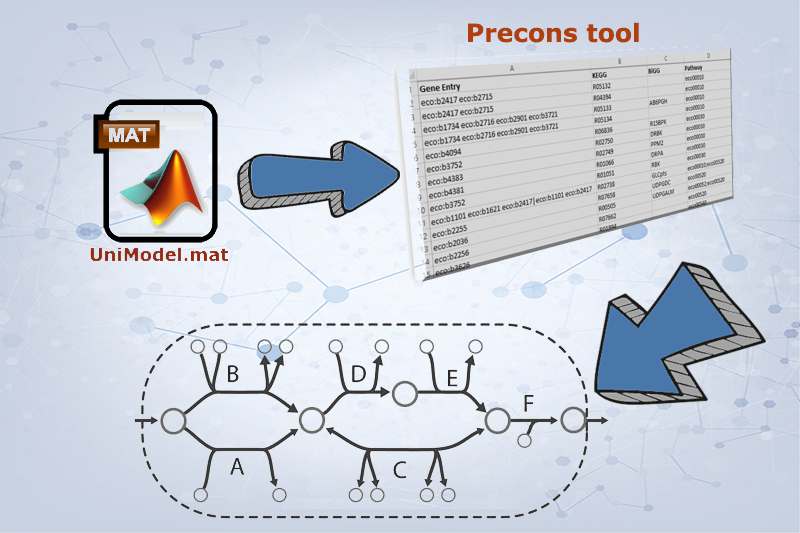
BiKEGG takes a flux distributions and a set of BiGG reaction IDs corresponding to the flux data (from COBRA toolbox) as input, and retrieve the set of equivalent KEGG reactions based on the previously generated KEGG reaction correspondences (e.g. UniModel). The user can then choose the set of KEGG pathways to be visualized. Finally, a set of equivalent KEGG reactions for the input BiGG model, the set flux data corresponding to this set of reactions and a number of users’ selected KEGG maps are generated. BiKEGG’s approach for visualization falls into two categories: 1- it can overlay the numerical results on the classical KEGG pathways; and 2- can generate customized metabolic maps based on KEGG global metabolic pathway. These customized metabolic maps contain reaction directionality based on BiGG models, which allow for a better understating and analysis of the visualized in silico results. Furthermore, BiKEGG is capable of visualizing the reaction fluxes for one or two different static conditions, and for dynamic conditions in an animated manner.